MoMo - Combining Neuron Morphology and Connectivity for Interactive Motif Analysis in Connectomes
Michael Shewarega -
Jakob Troidl -
Oliver Alvarado Rodriguez -
Mohammad Dindoost -
Philipp Harth -
Hannah Haberkern -
Johannes Stegmaier -
David Bader -
Hanspeter Pfister -
Download preprint PDF
Download Supplemental Material
Room: Hall E2
2025-11-06T14:45:00.000ZGMT-0600Change your timezone on the schedule page
2025-11-06T14:45:00.000Z
https://youtu.be/vzBq3yA2yUs
Keywords
Visual motif analysis, Scientific visualization, Neuroscience, Connectomics.
Abstract
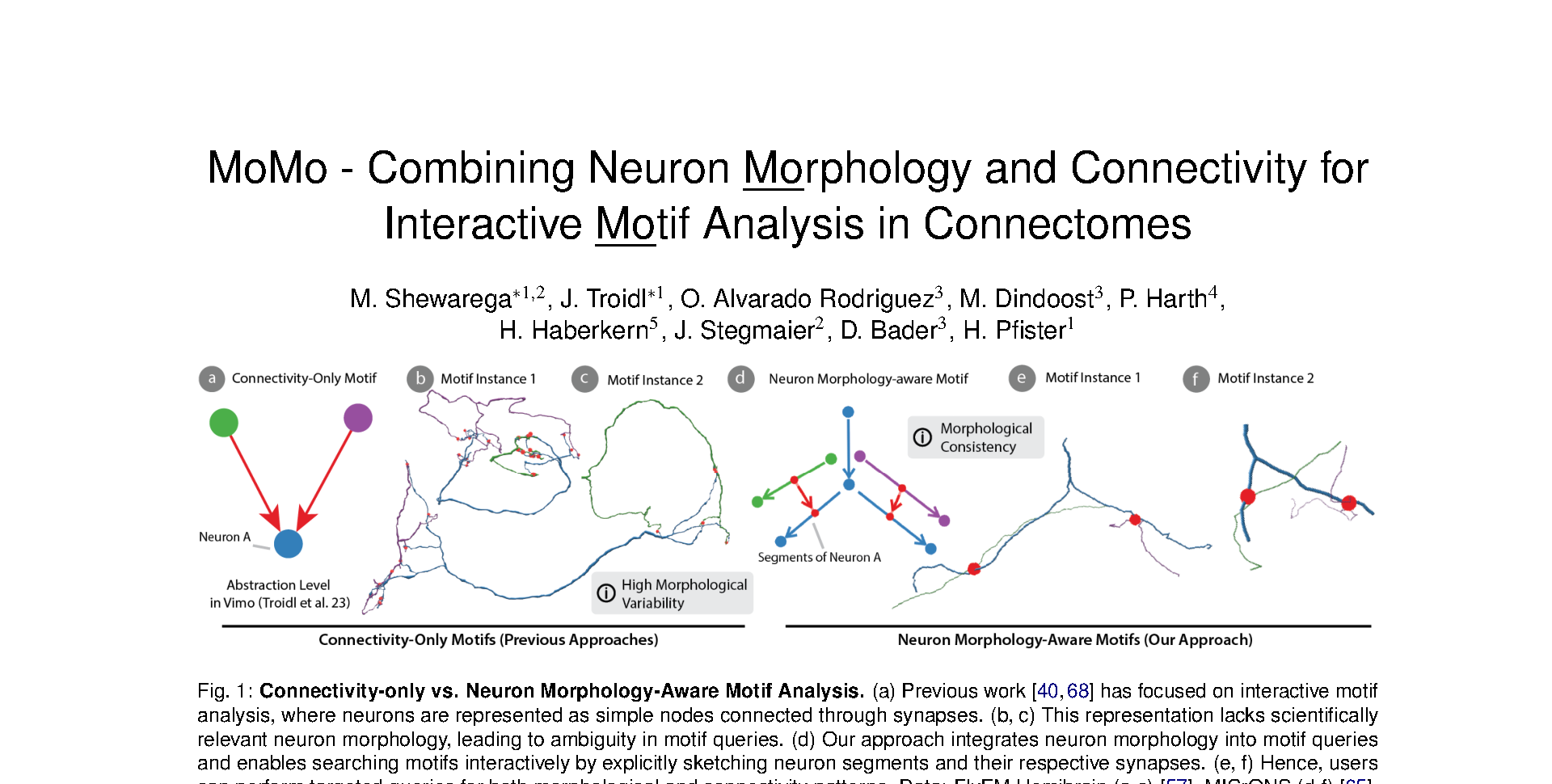
Connectomics, a subfield of neuroscience, reconstructs structural and functional brain maps at synapse-level resolution. These complex spatial maps consist of tree-like neurons interconnected by synapses. Motif analysis is a widely used method for identifying recurring subgraph patterns in connectomes. These motifs, thus, potentially represent fundamental units of information processing. However, existing computational tools often oversimplify neurons as mere nodes in a graph, disregarding their intricate morphologies. In this paper, we introduce MoMo, a novel interactive visualization framework for analyzing neuron morphology-aware motifs in large connectome graphs. First, we propose an advanced graph data structure that integrates both neuronal morphology and synaptic connectivity. This enables highly efficient, parallel subgraph isomorphism searches, allowing for interactive morphological motif queries. Second, we develop a sketch-based interface that facilitates the intuitive exploration of morphology-based motifs within our new data structure. Users can conduct interactive motif searches on state-of-the-art connectomes and visualize results as interactive 3D renderings. We present a detailed goal and task analysis for motif exploration in connectomes, incorporating neuron morphology. Finally, we evaluate MoMo through case studies with four domain experts, who asses the tool’s usefulness and effectiveness in motif exploration, and relevance to real-world neuroscience research. The source code for MoMo is available here: https://github.com/VCG/momo