Can LLMs Bridge Domain and Visualization? A Case Study on High-Dimension Data Visualization in Single-Cell Transcriptomics
Qianwen Wang -
Xinyi Liu -
Nils Gehlenborg -
Download preprint PDF
Download Supplemental Material
Room: Hall E1
2025-11-05T14:00:00.000ZGMT-0600Change your timezone on the schedule page
2025-11-05T14:00:00.000Z
https://youtu.be/zLchh59XyM4
Keywords
High dimensional visualization; LLM-supported literature review; Visualization in the wild
Abstract
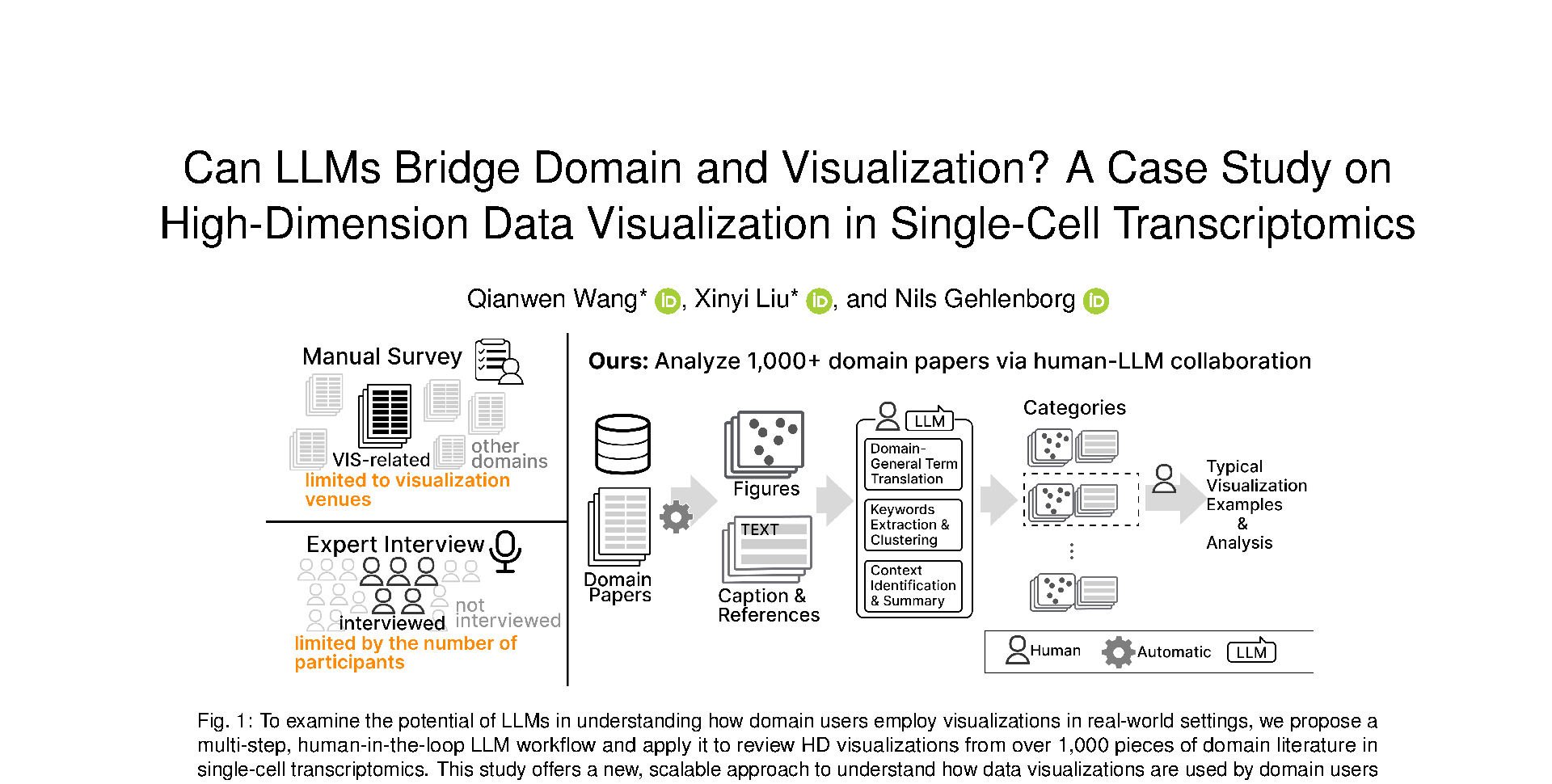
While many visualizations are build for domain users (e.g., biologists, machine learning developers), understanding how visualizations are used in the domain has long been a challenging task. Previous research has relied on either interviewing a limited number of domain users or reviewing relevant application papers in the visualization community, neither of which provides comprehensive insight into visualizations in the wild of a specific domain. This paper aims to fill this gap by examining the potential of using Large Language Models (LLM) to analyze visualization usage in domain literature. We use high-dimension (HD) data visualization in sing-cell transcriptomics as a test case, analyzing 1,203 papers that describe 2,056 HD visualizations with highly specialized domain terminologies (e.g., biomarkers, cell lineage). To facilitate this analysis, we introduce a multi-step, human-in-the-loop LLM workflow. Instead of relying solely on LLMs for end-to-end analysis, our workflow enhances analytical quality through 1) integrating image processing and traditional NLP methods to prepare well-structured inputs for three targeted LLM subtasks (i.e., translating domain terminology, summarizing analysis tasks, and performing categorization), and 2) establishing checkpoints for human involvement and validation throughout the process. The analysis results was validated with expert interviews and a test set, revealing three often overlooked aspects in HD visualization: trajectories in HD spaces, inter-cluster relationships, and dimension clustering. This research provides a stepping stone for future studies seeking to use LLMs to bridge the gap between visualization design and domain-specific usage.